Input data and parameters
Input
| Analysis date: | Fri Apr 25 11:09:09 EDT 2025 |
| BAM file: | /sc/arion/projects/BiNGS/bings_omics/data/bings/2025/bulk_course_data/bulkrna_pipeline/data_rna/preprocessed/star/Touchdome_1/Touchdome_1Aligned.sortedByCoord.out.bam |
| Counting algorithm: | uniquely-mapped-reads |
| GTF file: | /sc/arion/projects/BiNGS/bings_omics/data/bings/2025/bulk_course_data/bulkrna_pipeline/supporting_files/annotation/mus_musculus/grcm38_gencode_M25/data/gencode.vM25.basic.annotation.filtered.gtf |
| Paired-end sequencing: | no |
| Protocol: | strand-specific-forward |
| Sorting performed: | no |
Summary
Reads alignment
| Number of mapped reads: | 75,422,705 |
| Total number of alignments: | 81,259,438 |
| Number of secondary alignments: | 5,836,733 |
| Number of non-unique alignments: | 9,296,252 |
| Aligned to genes: | 50,937,525 |
| Ambiguous alignments: | 416,254 |
| No feature assigned: | 20,471,096 |
| Missing chromosome in annotation: | 138,311 |
| Not aligned: | 0 |
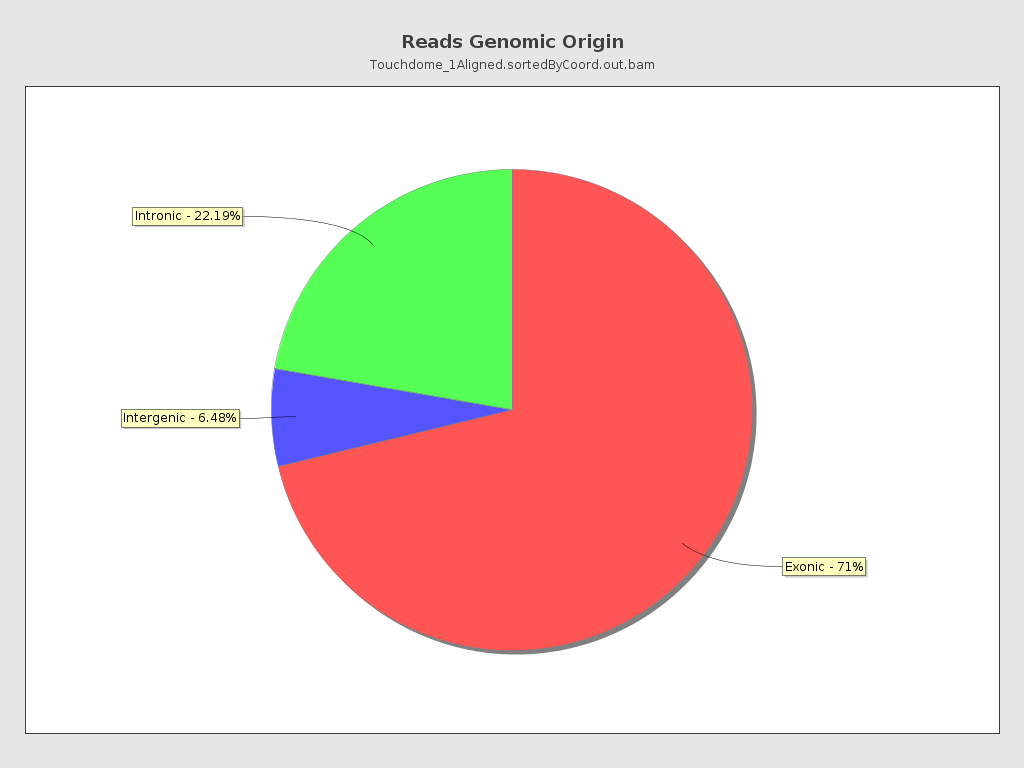
Reads genomic origin
| Exonic: | 50,937,525 / 71.33% |
| Intronic: | 15,845,065 / 22.19% |
| Intergenic: | 4,626,031 / 6.48% |
| Intronic/intergenic overlapping exon: | 5,412,065 / 7.58% |
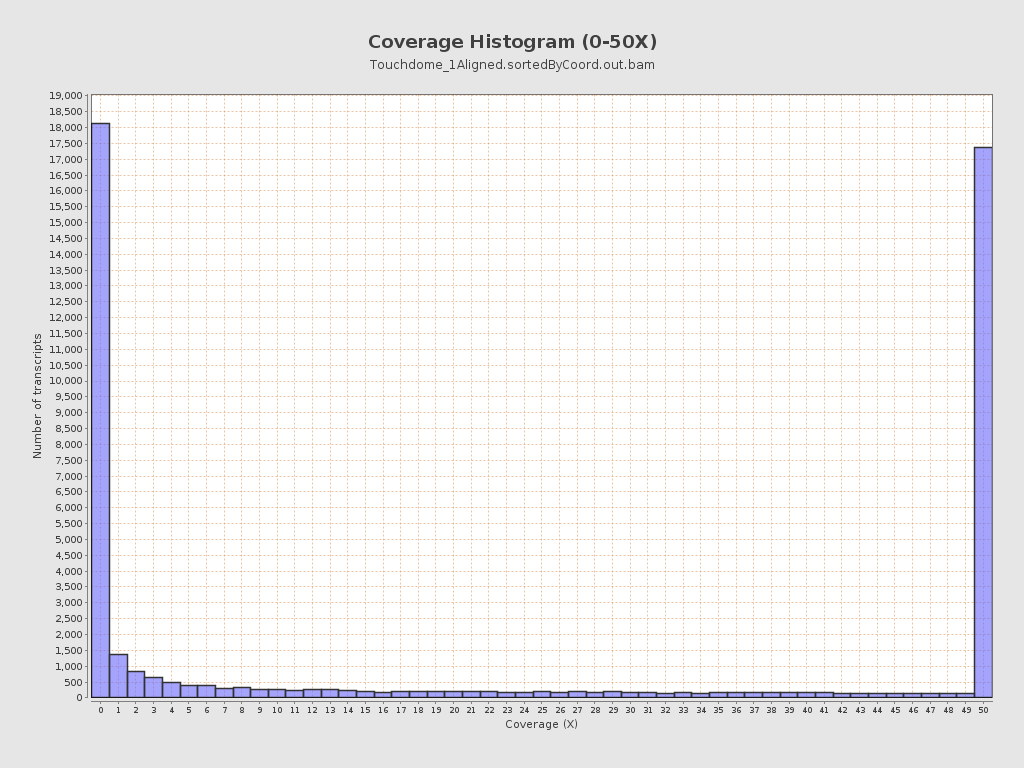
Transcript coverage profile
| 5' bias: | 0.76 |
| 3' bias: | 0.46 |
| 5'-3' bias: | 1.36 |
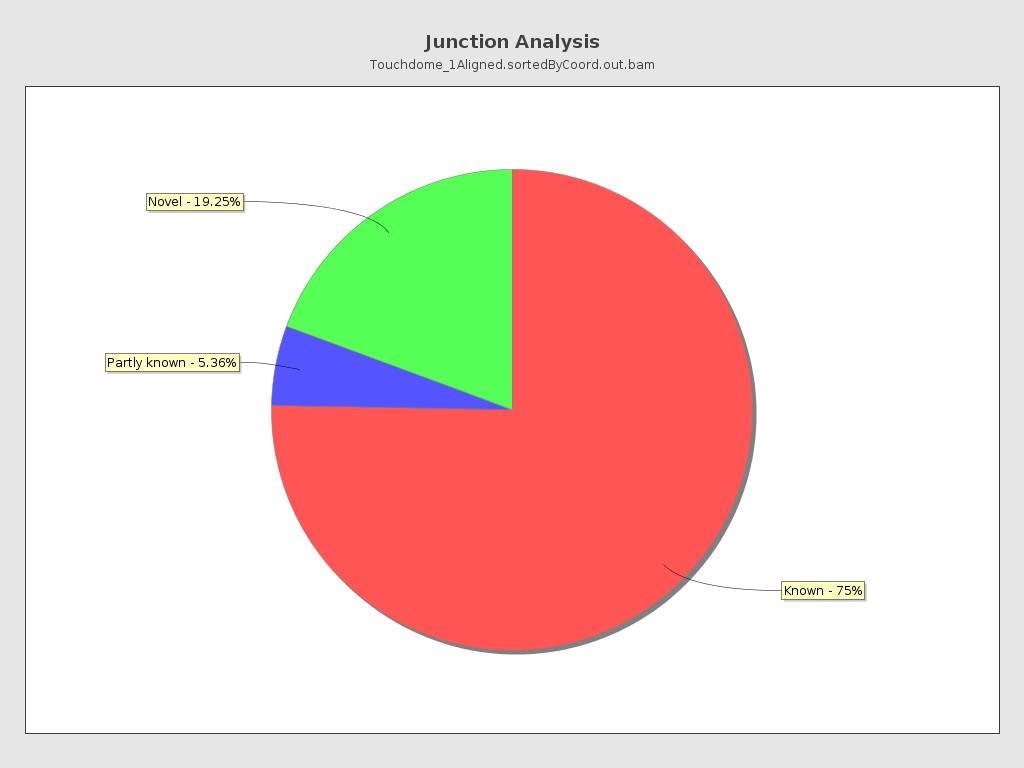
Junction analysis
| Reads at junctions: | 29,679,066 |
| ACCT | 5.35% |
| AGGT | 4.8% |
| TCCT | 4.65% |
| AGGA | 3.47% |
| GCCT | 2.99% |
| AGCT | 2.87% |
| ATCT | 2.83% |
| AGGC | 2.51% |
| AGGG | 2.45% |
| AGAT | 2.33% |
| CCCT | 2.26% |

.png)
.png)
.png)